14
- What is Microbial Biotechnology
- Types of Microorganisms
- What is Microbial Biotechnology
Learning Objectives
By the end of this section, you will be able to:
- Describe the different stages in the development of biotechnology
- Give examples of products that represent the different stages of biotechnology
Microbial biotechnology can be defined as the use of technology to improve or manipulate microorganisms or their products to generate products which are of benefit to man. In this book, the reader will learn about the different technologies which are used to manipulate microorganisms, the microbial products, and how modern genetic techniques are used to further improve the products. There are many applications of microbial biotechnology in diverse fields. This book will cover uses in forensics, agriculture, medicine, environmental protection, and many other industries.
The evolution of the science of biotechnology
The evolution of biotechnology as a science can be divided into two stages, classical and modern biotechnology, where modern mainly involves the use of recombinant DNA technology.

Figure 1. Winemaking in ancient Egypt is an example of classical biotechnology where microbes that naturally occur in nature were used to make a product for human consumption. Painting from the tomb of Nebamun, an Egyptian scribe and grain accountant from 1350 BC. https://commons.wikimedia.org/wiki/File:Nebamun_Supervising_Estate_Activities,_Tomb_of_Nebamun_MET_DT11772_detail-8.jpg
Classical biotechnology includes the most ancient form of biotechnology and that is when early humans took wild animals and plants from their natural habitat and started to grow them in conditions that they can better control; plants grown in soil that were tilled and fertilized and shepherded animals. That ancient form of biotechnology was followed by the selective breeding of plants and animals, where the male and female of a species that possess desirable traits were mated to produce offspring that would have the combined beneficial trait or traits of the parents. Fermentation is considered biotechnology and is a science that has been practiced for thousands of years in ancient Egypt (Fig. 1), Persia and China. Products of classical fermentation, which have used microbes found in nature, include wine or alcohol, bread, cheese, yogurt, vinegar, antibiotics, vaccines, vitamins, and many industrial enzymes (Fig. 2). Many antibiotics are derived from fungi and bacteria. Classical forms of vaccines include microbes that have simply been weakened or killed chemically by formaldehyde, or physical means using heat or irradiation. These vaccines include those against measles, mumps and rubella (MMR) and diphtheria, pertussis and tetanus (DPT). Many industrial enzymes used in making cheese, jellies, and detergents are from naturally occurring microbes. In agriculture, there are microbes used as biopesticides to control plant pests and diseases. The spores of Bacillus thuringiensis are effective against caterpillars, while Pseudomonas fluorescens is used to controlled fungal and bacterial diseases. Sewage treatment to get rid of environmental pollutants is another application of microbial biotechnology.

Figure 2. Products of classical microbial biotechnology include, clockwise from top left: Wine, yogurt, bread, and cheese (CC).
In contrast to classical biotechnology which makes use of microbes naturally isolated from nature, modern biotechnology employs recombinant DNA technology (RDT) to engineer microbes or improve their products. RDT is simply the linking together of at least two pieces of DNA from two different sources to come up with the hybrid, or recombined DNA. RDT is often use in genetic engineering which involves transfer genes from one genus to a different genus to enable the creation of genetically modified organisms (GMOs). This is in contrast to classical selective breeding where gene transfer can only happen successfully between closely related organisms, often within the same species. There are many examples now of modern fermentation where industrial strains have been genetically engineered to be able to produce increased amounts of industrial enzymes or metabolites such as amino acids or vitamins. A cheese-making enzyme, chymosin which used to be purified from calf intestines is now produced recombinantly in a fungus. Novel antibiotics not found in nature have been made by assembling biosynthetic pathway genes in microbes. RDT has been used to inactivate virulence genes in pathogenic microbes in the process of making them into vaccines. Genes from the human papilloma virus (HPV) has been transferred to yeast to make a vaccine against HPV. Human proteins like insulin, growth hormone, and other biotherapeutic proteins have been produced in E. coli or yeast. Heat tolerant or more catalytically active industrial enzymes like that used in oil drilling and even the textile industry have also been produced. In the field of agriculture, microbes have been engineered to be better at excluding pathogenic microbes. Microbial genes have also been engineered into plants to fight of insect pests and diseases. Animals have also been engineered to be faster growing or be less susceptible to viral infection.
1.2 Types of microorganisms
Learning Objectives
By the end of this section, you will be able to:
- List the major groups of microorganisms and give examples of each
- Describe the differences and similarities among the different types of microbes
- Describe the properties of Gram-positive and Gram-negative bacteria
- Explain Gram-staining
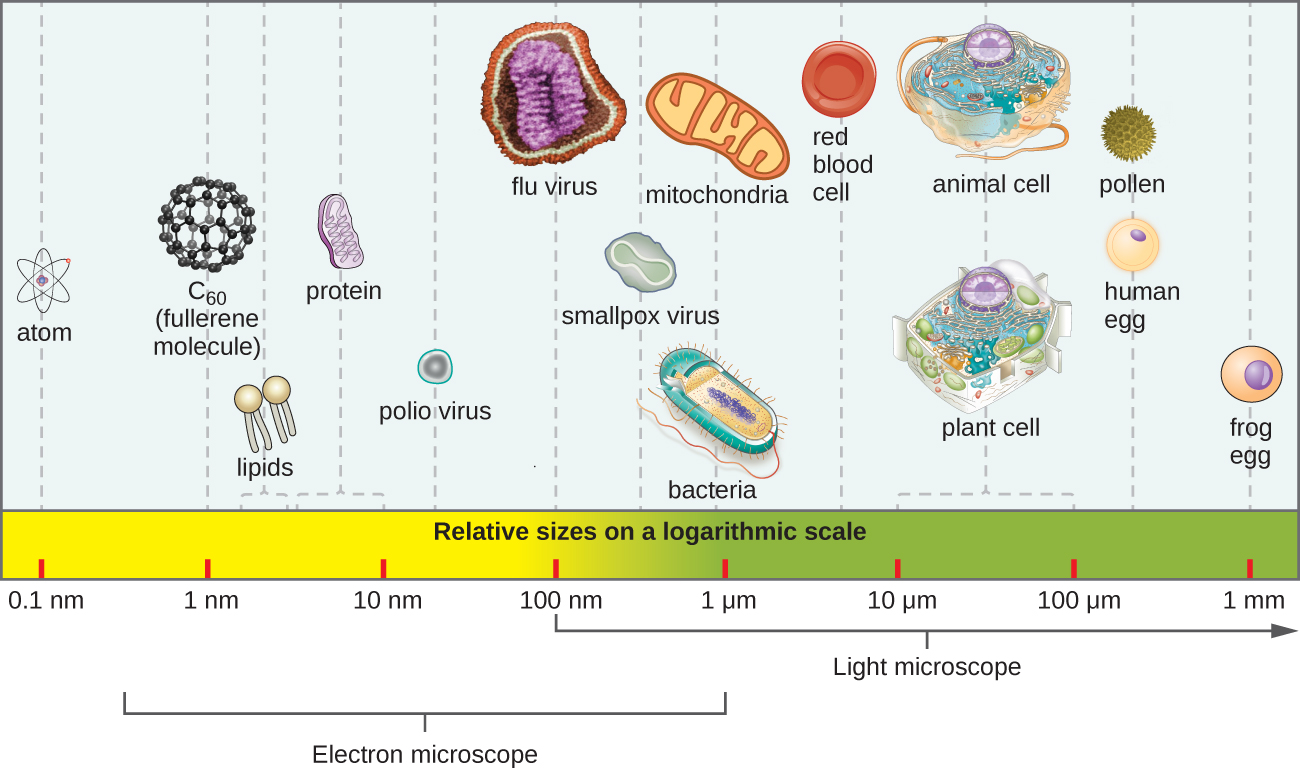
Figure 2. The relative sizes of various microscopic and non–microscopic objects. Note that a typical virus measures about 100 nm, 10 times smaller than a typical bacterium (~1 µm), which is at least 10 times smaller than a typical plant or animal cell (~10–100 µm). An object must measure about 100 µm to be visible without a microscope. https://cnx.org/contents/5CvTdmJL@9.36:eOZ_A6ye@13/1-3-Types-of-Microorganisms
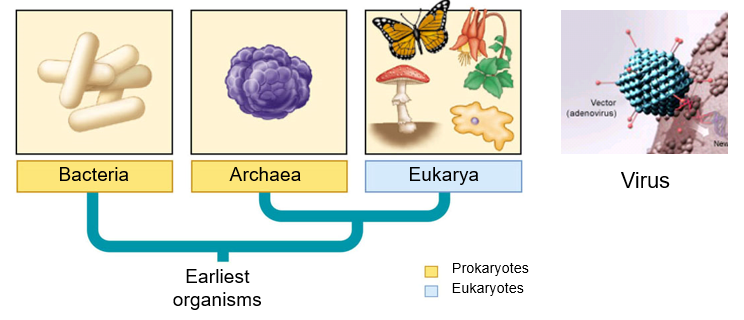
Figure 3. Microbes belong to the domains, bacteria, archaea, and eukarya. Bacteria and archaea are prokaryotes (yellow bars) having no membrane-bound organelles, while the eukarya is shaded blue. Viruses are included for their significance in medicine, both as causes and as treatment for diseases. Need new figure.
Microbes are defined simply as organisms for which one would need a microscope to be able to see them as individual cells are not visible with the naked eye (Fig. 2). Microbes have been around for three billion years; they have had that extensive period to evolve many capabilities to be able to successfully colonize and persist in every nook and cranny of the earth. A virus is not considered a true living organism because its metabolism or biological functions, including replication, are dependent on being able to infect a host cell. It can’t replicate on its own. However, viruses are included in this book for their significance as disease-causing agents in plants and animals, but more recently, for their use for the treatment of various diseases.
Microbes are classified under all three domains of living organisms: the bacteria, archaea and eukarya. Bacteria and archaea are prokaryotic in that they do not possess membrane-bound organelles, while eukaryotes have membrane-bound organelles, including the nucleus, for compartmentalization of certain cellular functions (Table 1). While the chromosomes in prokaryotes are not surrounded by a nuclear membrane, as in eukaryotes, it is concentrated in an irregularly shaped area of the cell called a nucleoid. When fully stretched, the length of the chromosome is about nine times that of the cell length. To be able to fit in the cell, it is compressed by supercoiling, which is facilitated by proteins. These proteins include histones in archaea. Peptidoglycan is present outside of the plasma membrane of bacteria, while absent in archaea and eukarya.
With exceptions, bacteria and archaea mostly have one chromosome while eukarya have two or more. Histones are associated with eukaryotic and archaeal chromosomes but are absent in bacteria. In bacteria, mRNAs can code for multiple protein products (polygenic), while that is rare in eukaryotes. In eukaryotes, non-protein coding regions of the mRNA, the introns, are common and spliced out during mRNA processing; introns are rare in bacteria and eukarya. To the 5’ end of mRNAs, methyl guanosine caps are added in eukaryotes, but not in bacteria and archaea; while a 3’ polyadenine tail is added to mRNAs in eukarya and archaea, but not in bacteria.
In both bacteria and archaea, the ribosomes consist of 30S small subunit and 50s large subunit, while the small and large subunits are 40S and 60S, respectively, in eukaryotes. The first amino acid incorporated during translation in archaea and eukarya is methionine, while it is formyl methionine in bacteria, although formyl methionine is also the initiator amino acid in the mitochondria and chloroplasts of eukaryotes.
In eukaryotes, transcription and mRNA processing takes place inside the nucleus; the mature mRNA is then translocated into the cytoplasm where the message is translated by ribosomes. In prokaryotes, transcription and translation both occur in the cytoplasm and translation can begin even before the mRNA is completely transcribed.
|
|
Bacteria |
Archaea |
Eukarya |
|
Membrane-bound organelles |
Absent |
Absent |
Present |
|
Nucleus |
Absent |
Absent |
Present |
|
Peptidoglycan |
Present |
Absent |
Absent |
|
|
|
|
|
|
Chromosome number |
1 |
1 |
>1 |
|
Polygenic mRNA |
Yes |
Yes |
None |
|
Introns |
Rare |
Rare |
Common |
|
5’ MeG cap |
No |
No |
Yes |
|
mRNA 3’ polyA |
None |
Yes |
Yes |
|
Ribosome |
30S, 50S = 70S |
30S, 50S |
40S, 60S = 80S |
|
Initiator tRNA |
Formyl methionine |
Methionine |
Methionine |
|
|
|
|
|
Table 1. Differences among microbial members of the three organismal domains.
Figure _________ A typical prokaryotic cell contains a cell membrane, chromosomal DNA that is concentrated in a nucleoid, ribosomes, and a cell wall containing peptidogycan. Some prokaryotic cells may also possess flagella, pili, fimbriae, and capsules. Adapted from Microbiology_OpenStaxCC. https://cnx.org/contents/5CvTdmJL@9.36:W1kvc5fi@15/3-3-Unique-Characteristics-of-Prokaryotic-Cells
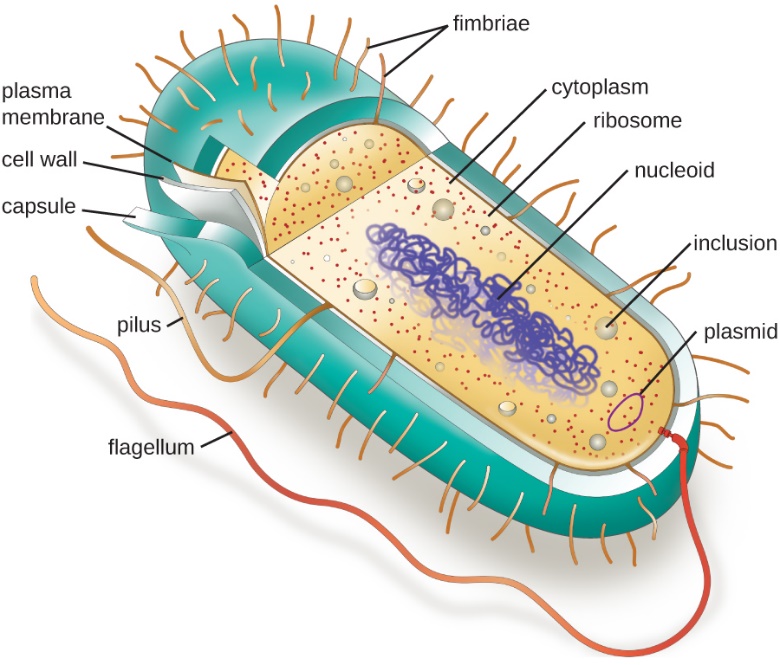
|
|
|
Figure ______. Gram positive (left) and Gram negative (right) bacteria. Joe Rubin. CC. FlickR. https://www.flickr.com/photos/therubinlab/50044882852 and B. Domangue https://commons.wikimedia.org/wiki/File:Shigella_flexneri_Gram_Stain_on_Microscope_Slide.jpg
|
Bacteria subgroups |
Gram + |
Gram – |
|
Gram stain |
Purple |
Red |
|
Peptidoglycan |
Thick |
Thin |
|
Cell membrane |
One |
Two |
|
|
|
|
|
Lipopolysaccharides |
Absence |
Present |
|
Representative genera |
Lactobacillus |
Escherichia |
|
|
Clostridium |
Pseudomonas |
|
|
Streptococcus |
Agrobacterium |
|
|
Streptomyces |
Yersinia |
|
|
Staphylococcus |
Rhizobium |
Table 2. Differences between Gram (+) and Gram (–) bacteria and some examples of each groups which will be mentioned later in this book.
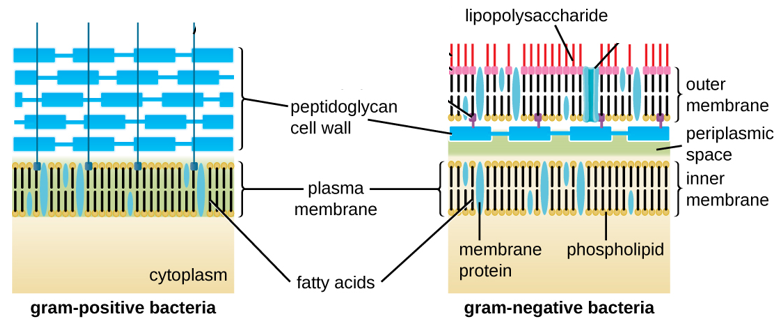
Figure ________. Bacteria contain two common cell wall structural types. Gram-positive cell walls are structurally simple, containing a thick layer of peptidoglycan external to the plasma membrane. Gram-negative cell walls are structurally more complex, containing three layers: the inner membrane, a thin layer of peptidoglycan, and an outer membrane containing lipopolysaccharide. (Credit: Modification of work by “Franciscosp2”/Wikimedia Commons).
https://commons.wikimedia.org/wiki/File:Bacteria_cell_wall.svg
(PARKER, SCHNEEGURT, THI TU, FORESTER and LISTER)
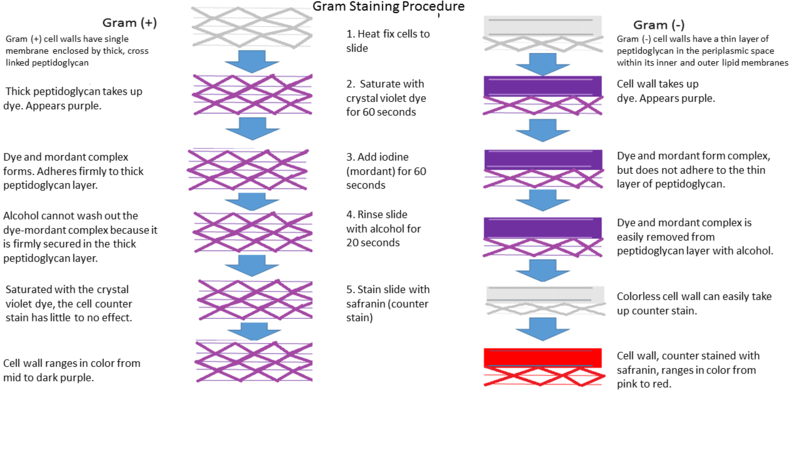
Figure ______. Gram staining steps in Gram positive and Gram negative bacteria. CC by Paityn Donaldson. https://commons.wikimedia.org/wiki/File:Gram_Stain.png
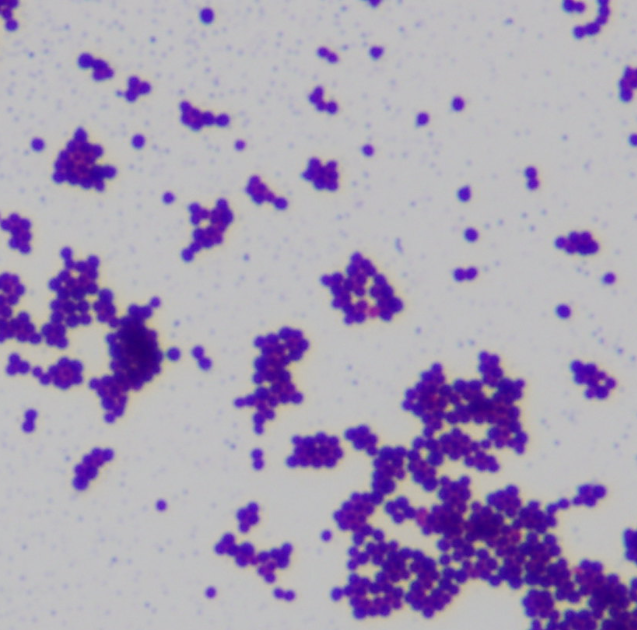
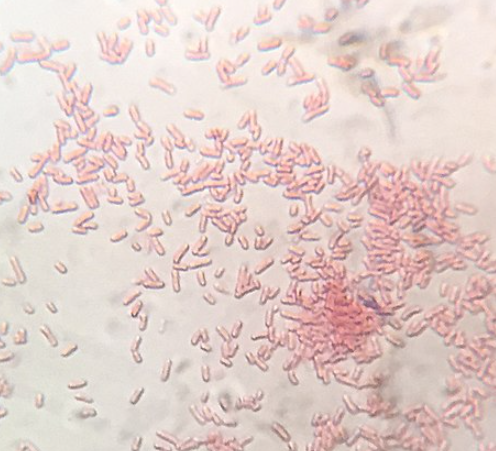
Bacteria are divided into two groups based on their cell wall structure as determined by their Gram staining properties (Table 2 and Fig ___). The Gram stain was developed by Hans Christian Gram in 1884 and is widely used to quickly diagnose unknown disease agents. It is also a standard test performed to identify bacterial contaminants in drug products and manufacturing facilities.
Gram-positive and Gram-negative bacteria stain purple and pink, respectively, with the Gram stain. The ability of Gram-positive bacteria to retain the crystal violet dye is due to the much thicker peptidoglycan layer in their cell wall (90% of cell wall), as compared to the thinner layer present in Gram negatives (10% of cell wall). Gram positive bacteria have a single cell membrane consisting of lipid bilayers, while Gram negative have an inner and a second outer membrane. On the outer cell membrane of Gram-negative bacteria are anchored lipopolysaccharides, also known as endotoxins, which can cause uncontrolled immune responses in humans and can lead to lethal consequences. A thick, gel-like periplasm is found in between the inner and outer cell membranes of Gram-negatives, while a much thinner periplasm is found between the cell membrane and peptidoglycan layer of Gram positives. Many bacteria also possess a thick, outermost layer of polysaccharide called the capsule. The capsule and lipopolysaccharides help protect bacteria against phage infections and host defense mechanisms.
In the Gram stain, the cells are first heat fixed, then stained with crystal violet, which is taken up in similar amounts by all bacteria. The cells are then treated with an iodine mordant to fix the stain, washed briefly with 95% alcohol (destained), and finally counterstained with safranin red. Gram-positive organisms retain the initial violet stain, while gram-negative organisms are decolorized by the organic solvent and hence show the pink counterstain. Some bacteria are gram-variable (that means, they may stain either negative or positive) while some bacteria are not susceptible to either stain used in the Gram technique.
Cell division. When prokaryotes (bacteria and archaea) attain a certain size, they begin to divide by a process called binary fission (Fig. __) . The DNA localizes near the center of cells, starts to undergo replication, and then the daughter strands separate. A cell wall and plasma membrane then form around the center of the cell. Upon cell wall completion, the daughter cells then separate, with each daughter cell carrying a copy of the bacterial genome. Note that the genetic material needs to be duplicated first or else cell division will not proceed. In eukaryotes, there is mitotic cell division which retains the original chromosome number of the parent cell and meiosis which leads to halving of the chromosome number.
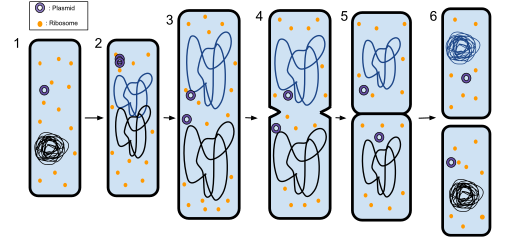
Figure ______. Prokaryotic cell division by binary fission. 1: The bacterium before binary fission has the DNA tightly coiled. 2: The cell elongates to the size of the parent cell and the DNA of the bacterium gets repllicated . 3: The DNA is pulled to the separate poles of the bacterium. 4: The growth of a new cell wall begins the separation of the bacterium. 5: The new cell wall fully develops, resulting in the complete split of the bacterium. 6: The new daughter cells have tightly coiled DNA, ribosomes, and plasmids. Modified from Ecoddington14. https://open.oregonstate.education/microbiology/chapter/3-3unique-characteristics-of-prokaryotic-cells/https://commons.wikimedia.org/wiki/File:Binary_Fission_2.svg
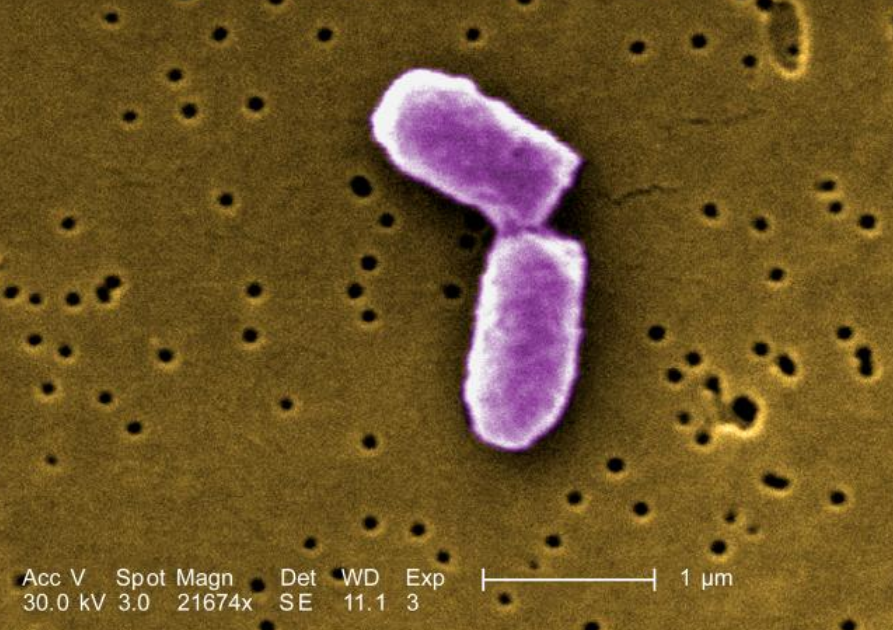
Fig. __. Digitally-colored, scanning electron microscopic (SEM) image of E. coli at a late stage of cell division where the cell wall has separated the two daughter cells. https://phil.cdc.gov/Details.aspx?pid=10042
Summary
- Microbial biotechnology:
- Is the use of technology to improve microbes for the benefit of man.
- Modern biotechnology makes use of recombinant DNA technology, while classical biotechnology does not
- Microorganisms belong to Bacteria, Archaea, and Eukaryotic groups.
- Bacteria can be grouped into Gram-positive and Gram-negative
- Some bacteria are coccoid, rod-shaped or spiral shaped
.